Our lab focuses on the development of new technologies for molecular analysis and biomedical research via advances in micro- and nano-scale sciences. We aim to develop new methods, devices and systems with unprecedented performance characteristics, such as sensitivity, specificity, resolution (temporal and/or spatial), multiplexing and throughput to rectify current technological limitations in the molecular study of diseases. In addition, we are moving beyond basic research into translational studies by developing and applying new technologies that address practical biomedical problems and clinical needs. By forging long-term collaborations with medical scientists and physicians across various disciplines (e.g. Oncology, Pathology, Surgery and Emergency Medicine) and by leveraging our engineering innovations in microfluidics, single molecule spectroscopy and functional nanoparticles, we are developing genetic and epigenetic biomarker-based diagnostics for cancer and an array of other diseases.
 Quantum Dots for Genetic and Epigenetic Detection of Cancer
Quantum Dots for Genetic and Epigenetic Detection of Cancer
Semiconductor nanocrystals, also known as quantum dots (QDs) have several superior photophysical properties compared to organic fluorophores, including wide absorption bands, size-tunable and narrow emission spectra, high brightness and photostability. By taking advantage of these properties, we developed a DNA nanosensor that detects target sequences via DNA hybridization and fluorescence resonance energy transfer (FRET) using QDs as energy donors. QDs are ideal donors in a FRET system that overcomes several difficulties encountered in traditional analysis of FRET signals, such as photobleaching, spectral cross-talk, and direct acceptor excitation. As a result, the QD-FRET nanosensor presented extremely low background but produced strong FRET signals upon binding to even a small amount of target DNA (~50 copies or less).
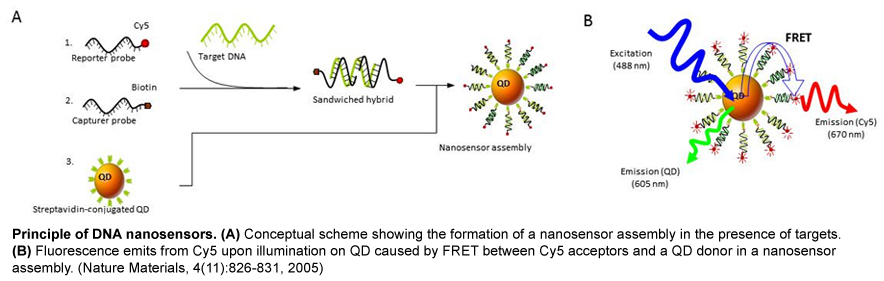
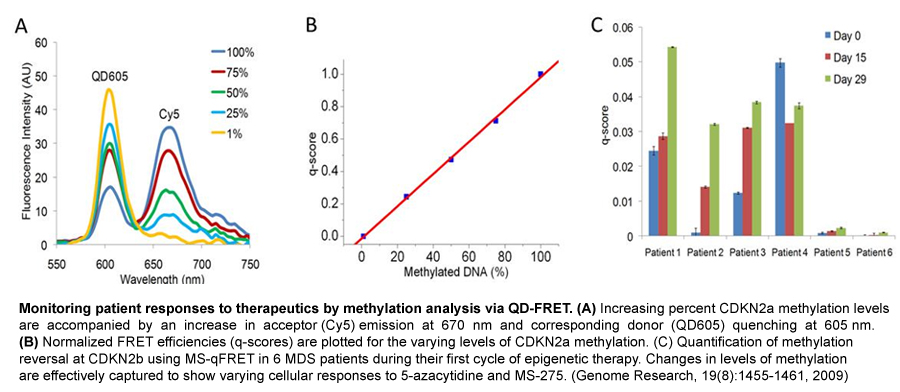
We have extended QD nanosensors for genetic detection of cancer by combining techniques of QD-FRET detection and DNA ligation and successfully demonstrated detection of Kras and Braf point mutations in ovarian cancer patient samples. We have further applied QD nanosensors to analyze a promising epigenetic biomarker of cancer DNA methylation. Aberrant DNA hypermethylation is observed at classic tumor-suppressor genes, which are known to be genetically mutated and cause inherited forms of cancer. Tumor cells display a larger number of genes inactivated by promoter hypermethylation than by genetic mutations. Furthermore, these abnormal epigenetic changes appear to be an early event that precedes detection of genetic mutations. Thus, detection of promoter hypermethylation is a valuable tool for early diagnosis of cancer, monitoring tumor behavior, as well as measuring response of tumors to targeted therapy. Our method was accomplished by integrating sodium bisulfite DNA conversion with QD-FRET probe designs. This epigenetic nanosensor boasts a remarkably high sensitivity for the detection of promoter DNA methylation, surpassing the current gold standard method: methylation specific PCR (MSP). We have demonstrated that the high sensitivity of the DNA nanosensor enables the direct screening of non-invasive clinical samples (sputum, stool and blood) for tumor-derived methylated DNA. Furthermore, this technique has shown great potential for the early detection of cancer, by demonstrating its effectiveness in the detection of cancer precursors and stage I cancer patients (lung and colon cancer).
 Single Molecule Analysis of Genomic Content
Single Molecule Analysis of Genomic Content
 All-in-One Microfluidic Diagnostics: Sample Preparation, Target Detection and Integration
All-in-One Microfluidic Diagnostics: Sample Preparation, Target Detection and Integration
 Droplet Microfluidics for High-Resolution and High-Throughput Screening
Droplet Microfluidics for High-Resolution and High-Throughput Screening